前面,我们在差异表达结果中按照padj筛选出了TOP10基因,现在,我们对TOP10基因绘制生存曲线,看它们的表达是否与病例的生存时间相关。
1 | > library(survival) |
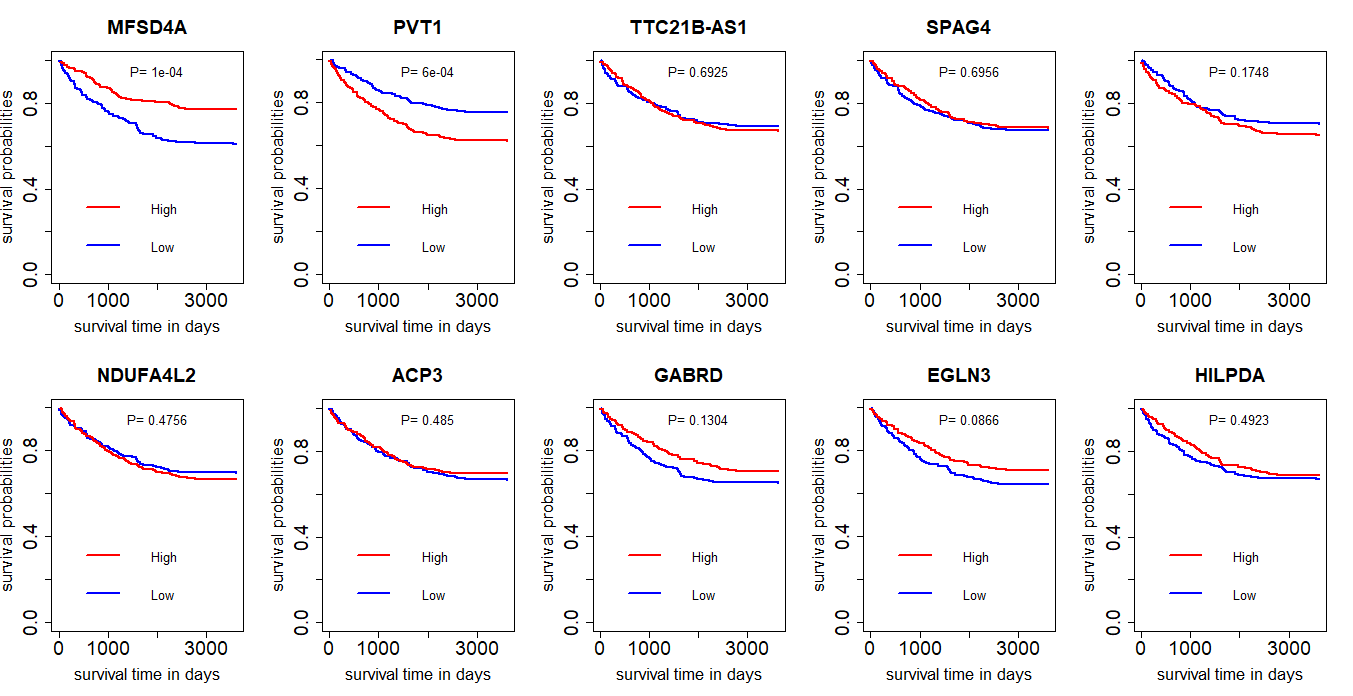
这个结果并不理想,只有前两个基因的表达量与生存时间有关联,MFSD4A高表达生存时间长,而PVT1低表达生存时间长。
我们可以换个角度思考同一个问题,如果以FoldChange来挑选TOP10基因,结果如何?
1 | > annot_order_FC <- result_select_annot[order(abs(result_select_annot$log2FoldChange),decreasing = T),] |
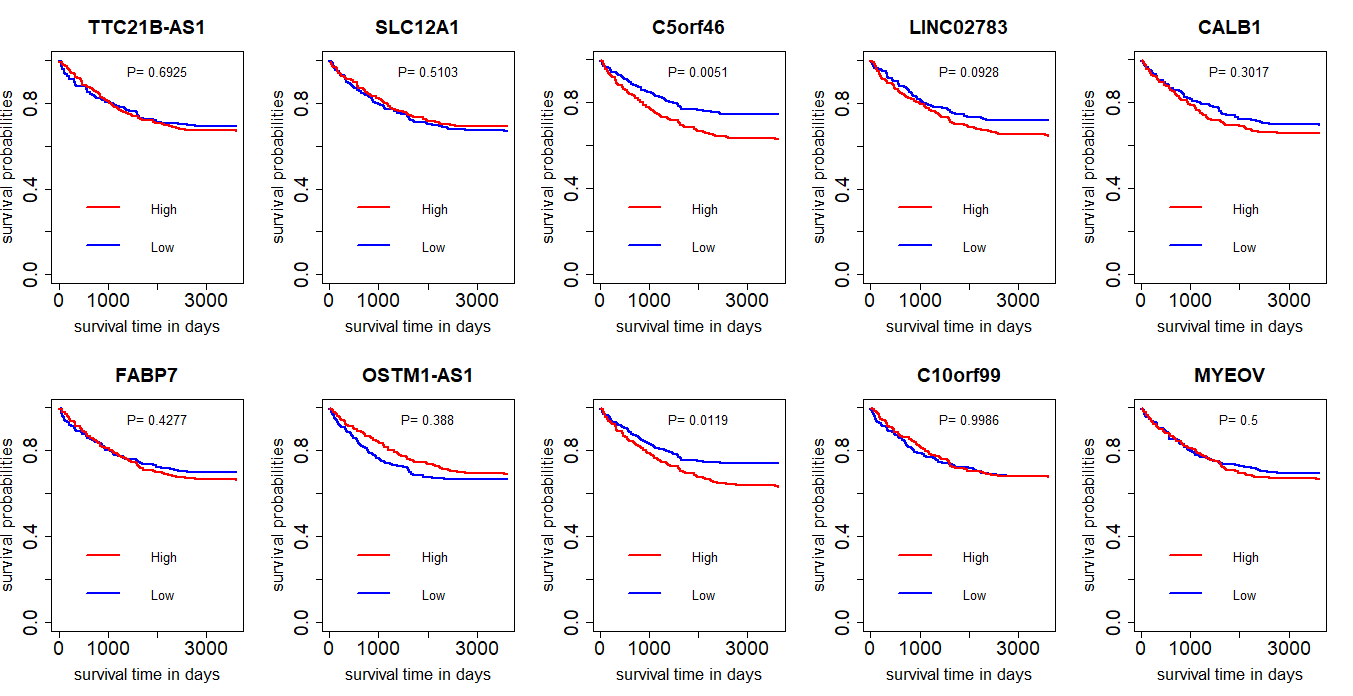
结果还不如使用padj,可见,表达差异大的基因不一定与生存时间有关联。
