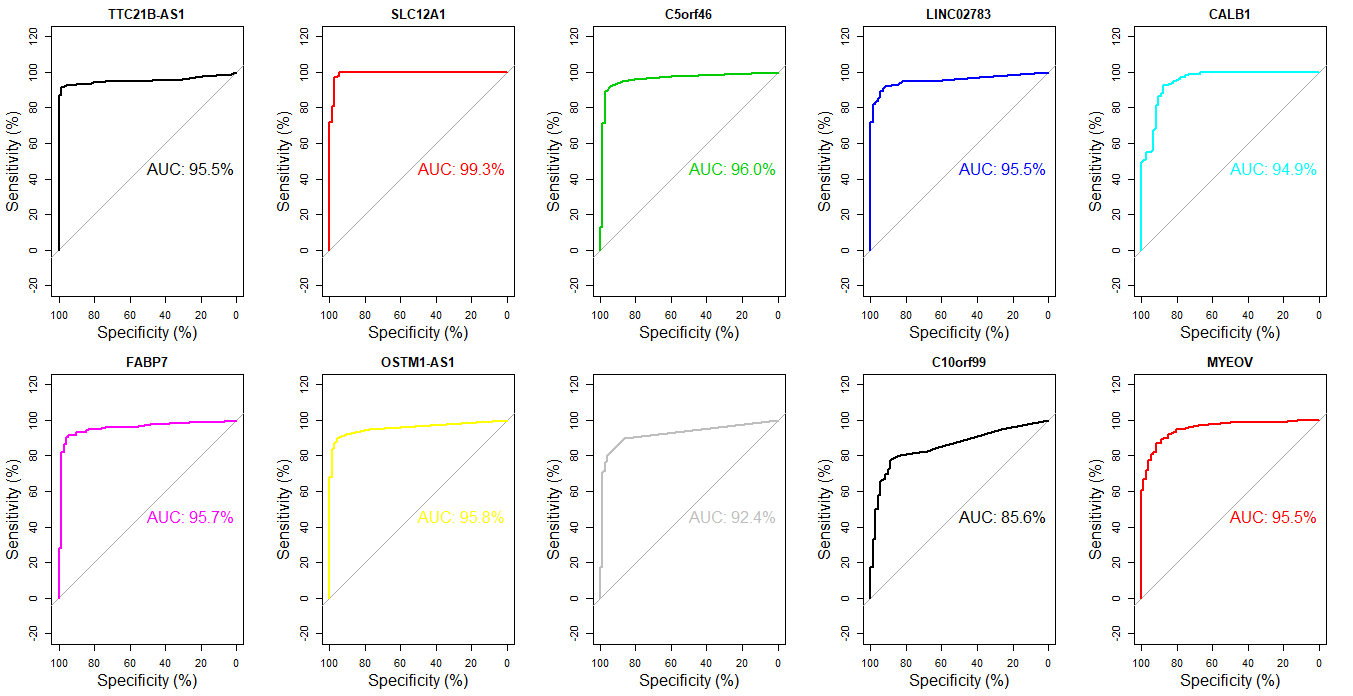
得到差异表达结果之后,我们可以根据padj来挑选排名前10的基因,来绘制受试者工作曲线,评估这些基因是否能作为判断肿瘤的标志基因。
1 | > annot_order <- result_select_annot[order(result_select_annot$padj),] # 按padj由小到大排序 |
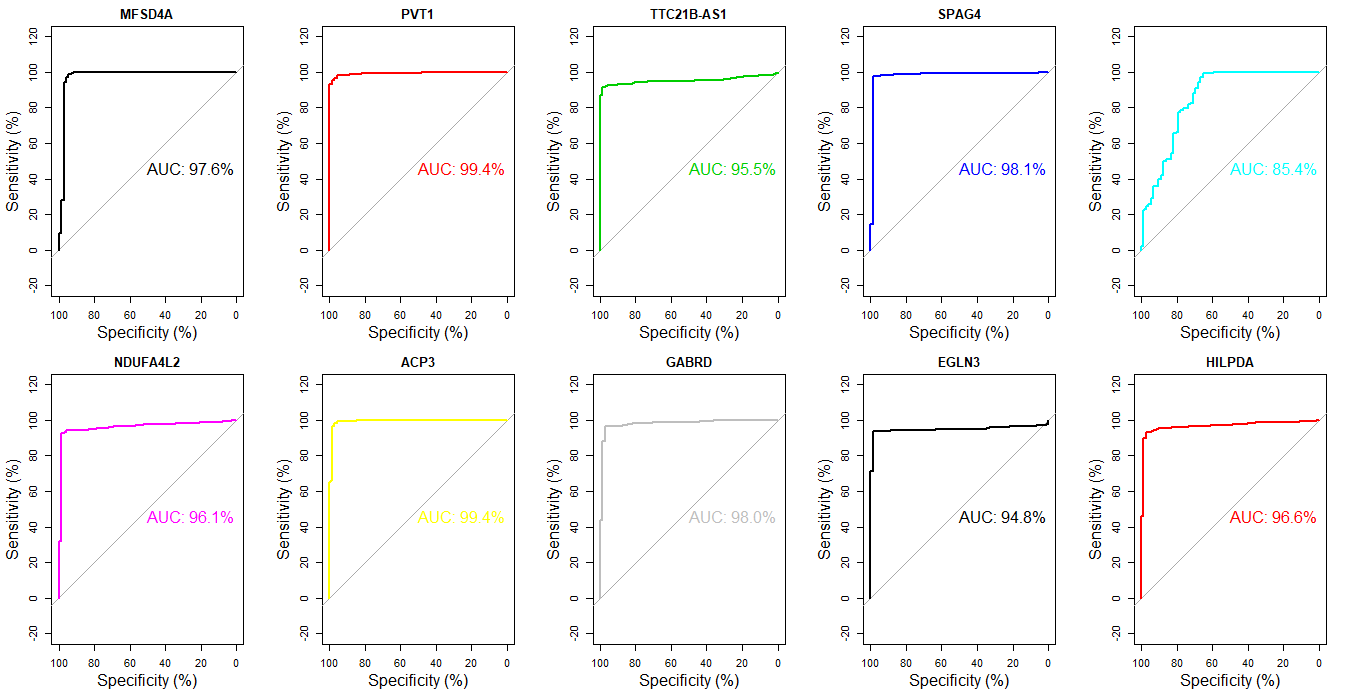
换个角度,如果以FoldChange筛选排名前10的基因,结果如何?
1 | annot_order_FC <- result_select_annot[order(abs(result_select_annot$log2FoldChange),decreasing = T),] |