Firehose是由Broad institute开发的,提供经过预处理后的TCGA数据,在R语言中,RTCGAToolbox可用于查询、下载和分析Firehose的数据。
1 | if (!requireNamespace("BiocManager")) |
在通过Firehose获取数据之前,需要检查有效的数据集,“getFirehoseDatasets”可以列出所有的数据集,而“getFirehoseRunningDates”与“getFirehoseAnalyzeDates”给出数据库更新日期。
1 | > getFirehoseDatasets() |
在确定数据集名称与更新日期后,就可以使用“getFirehoseData()”来获取数据了,下面来下载数据,以READ(直肠腺癌)为例:
1 | > brcaData <- getFirehoseData(dataset="READ", runDate="20160128", |
对于getFirehoseData(),以下几个参数是必选的:
1)dataset,可以通过getFirehoseDatasets() 获取;
2)tunData,可以通过getFirehoseRunningDates()获取;
3)gistic2Date,如要获取copy number data,则必须设定这个参数,可以通过getFirehoseAnalyzeDates()获取;
可选参数如下:
1)forceDownload,是否强制下载或使用工作目录中以前下载的旧数据;
2)fileSizeLimit,默认为500MB;
3)getUUIDs,获取UUIDs条形码;
除Clinical之外,还有如下参数可选:
RNAseqGene
clinical
miRNASeqGene
RNAseq2GeneNorm
CNASNP
CNVSNP
CNASeq
CNACGH
Methylation
Mutation
mRNAArray
miRNAArray
RPPAArray
下载完成后,可以使用biocExtract()来提取数据,如提取clinical data,
1 | > clinicData=biocExtract(brcaData,'clinical') |
下面提取mutation data
1 | > mutationData = biocExtract(brcaData,'Mutation') |
下面,可以先看一下突变情况
1 | > mt=getMutationRate(brcaData) |
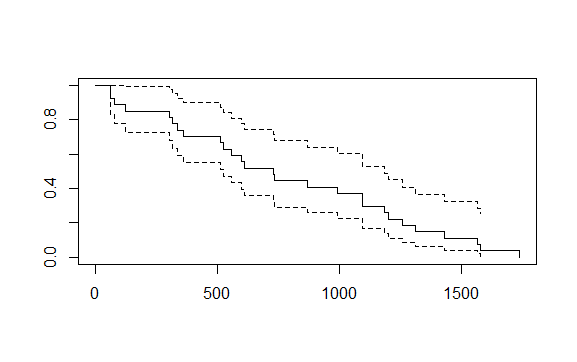
绘制生存曲线
1 | > survData <- data.frame(Samples=rownames(clinicData), |