一、加载程序包
1 | library(survival) |
二、加载数据并绘图
1 | data <- read.delim('clipboard',header = T) |
1 | head(data) |
1 | table(data$Status) |
1 | Alive Dead |
1 | table(data$Group,data$Status) |
1 | Alive Dead |
1 | my.surv <- Surv(data$Days,data$Status=='Dead') |
1 | fit <- survfit(my.surv~data$Group) |
1 | Call: survfit(formula = my.surv ~ data$Group) |
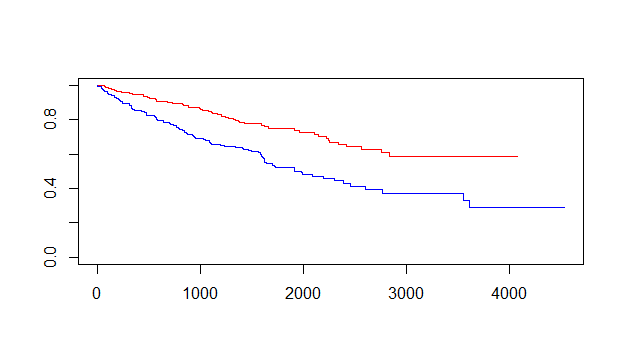
绘制最基本的生存曲线1
plot(fit,col=c("red","blue"))
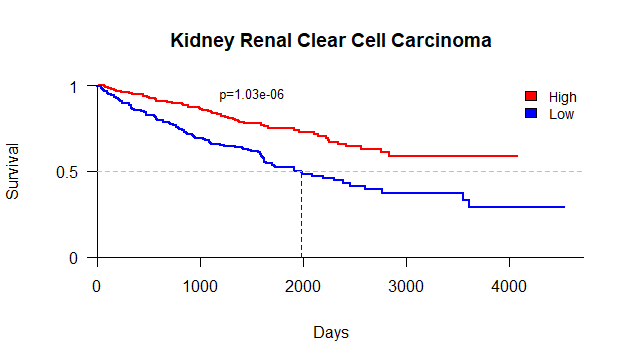
优化生存曲线
1 | plot(kmfit2,col=c("red","blue"),axes=F,xlab="Days",ylab="Survival",lwd=2,main="Kidney Renal Clear Cell Carcinoma") |